.jpg)
Research programmes in grapevine and wine biotechnology Introduction The IWBT is the only research institute in the country that is focused primarily on studying the biology of grapevine and wine microorganisms, and cooperates very closely with the wine and table grape industries of South Africa. Approach to research The research philosophy of the IWBT is the result of strategic decisions on how best to respond to five challenges:
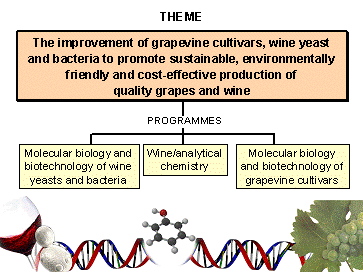
Research portfolio The IWBT's research theme is the understanding of the biology of wine-associated organisms, including the ecology, physiology, molecular and cellular biology of grapevine, wine yeast and wine bacteria to promote the sustainable, environmentally friendly and cost-effective production of quality grapes and wine. The Institute continually integrates the latest technologies in the biological, chemical, molecular and data analytical sciences to achieve these aims. The specific research portfolio consists of three programmes (see Fig. 1). The first focuses on a better understanding and exploitation of wine associated microbial biodiversity, and the physiological, cellular and molecular characterisation of Saccharomyces and non-Sacharomyces yeasts, as well as the genetic improvement of wine yeast strains. A second programme is concerned with lactic acid and other bacteria, including their impact on wine, metabolic characterisation and improvement of malolactic fermentation. The third programme focuses on the physiology, cellular and molecular biology and genetic improvement of grape cultivars. These programmes are planned and executed in an integrated manner, combining research groups from the Department of Chemistry and Polymer Science, the Department of Food Science, the Department of Viticulture and Oenology and the Institute for Wine Biotechnology under the scientific theme of Metabolomics and Metrics of vine, wine and wine organisms. The focus of the research will be to provide a fully integrated and controlled research chain starting with characterised model vineyards and ending with a comprehensive chemical, sensory and quality assessment of the final product. Each of these partners offers specific postgraduate opportunities
Grapevine molecular biology and biotechnology Dr John Moore
Prof Melané Vivier
Dr Philip Young
Wine biotechnology Prof Florian Bauer
Dr Benoit Divol
Prof Maret du Toit
Dr Evodia Setati
Computational Biology and Biomathematics Dr Dan Jacobson
Analytical chemistry Dr Helénè Nieuwoudt
The IWBT's research is conducted in collaboration with local research institutions, as well as several international laboratories. National
International
Scientific articles 2013
| |||||||||||||||||||||||||||||||||||||||||||||||
PAGEMASTER: Tel: +27-21-808 3770; Fax: +27-21-808 3771; E-mail:
Pagemaster
| |||||||||||||||||||||||||||||||||||||||||||||||